Bioinformatics research
Bioinformatics studies of microorganisms, in particular actinomycetes, occupy an important place in modern microbiology, genetics and molecular biology. Thanks to the development of high-throughput genome sequencing technologies and the constant increase in the volume of genetic data, bioinformatics has become a powerful tool for understanding the functions and evolutionary relationships between individual genes and even different species of organisms.
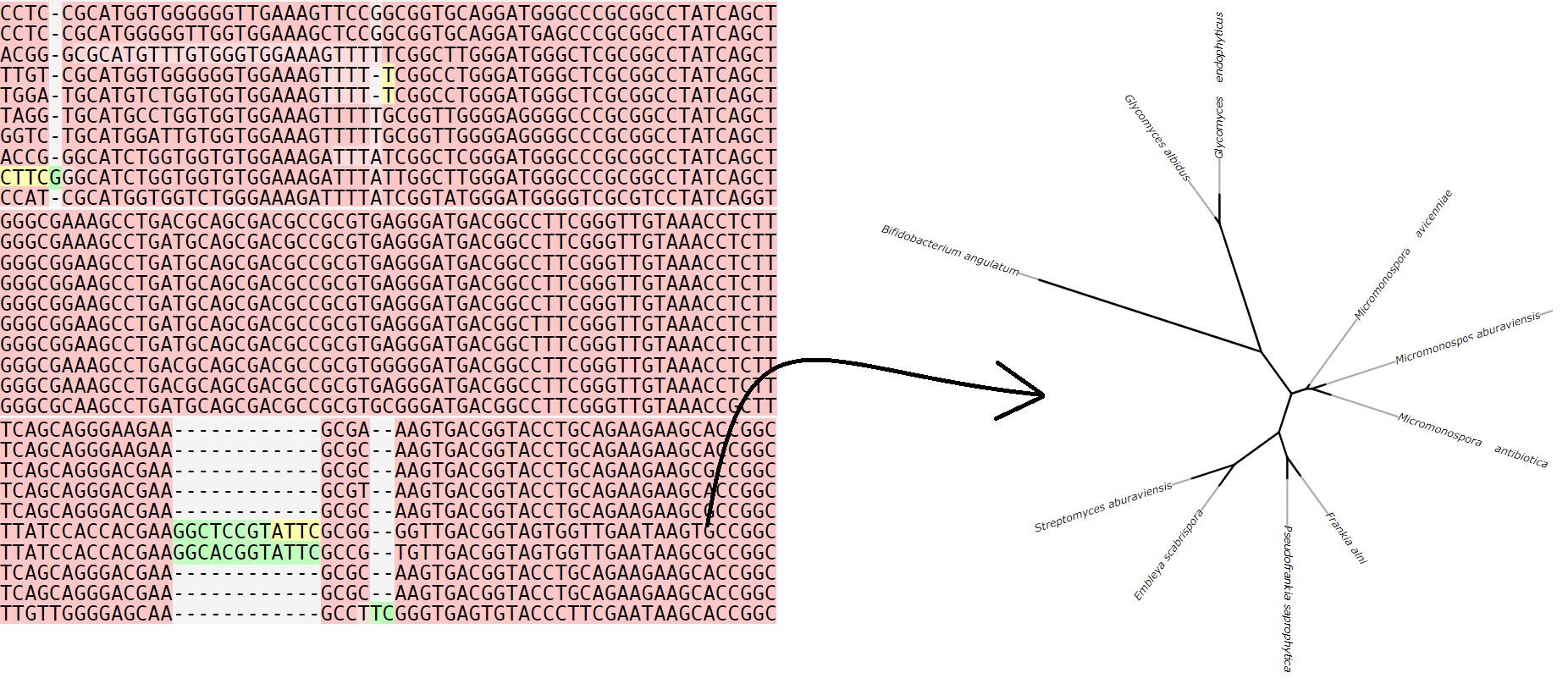
Our team is actively investigating individual genes, especially the 16S rRNA gene, which is a common approach for determining phylogenetic relationships between organisms. The 16S rRNA gene is a particle of the ribosome that has a conservative structure, but at the same time has sufficiently variable regions that are quite universal markers for establishing the systematic position of the bacteria under study. We use the obtained nucleotide sequences to build phylogenetic trees, which are a graphic representation of the diversity and evolutionary relationships between natural isolates of microorganisms.
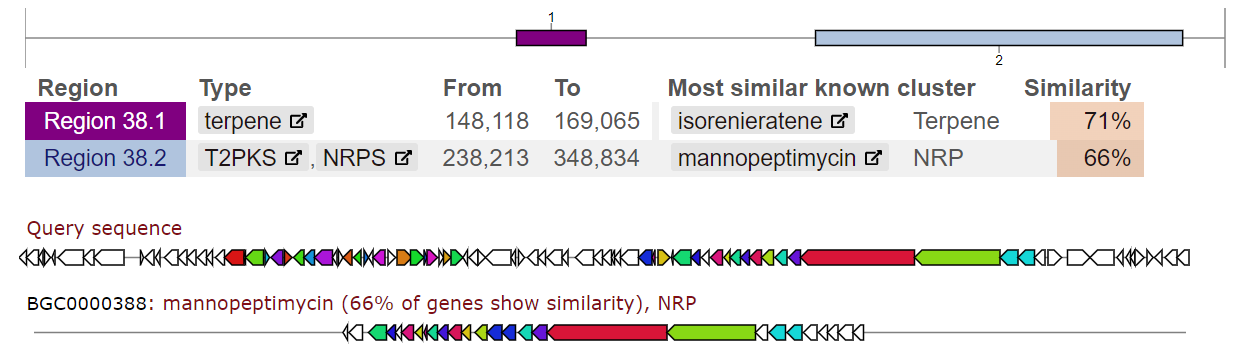
With the development of sequencing technologies, the analysis of whole genomes has become common. By sequencing and assembling genetic material, we obtain complete nucleotide sequences of bacterial genomes, identify individual genes responsible for important biological functions, and screen biosynthetic gene clusters encoding potentially new biologically active compounds. Annotation of genomes and the search for clusters of secondary compounds are important stages of bioinformatics research. These methods make it possible to obtain information about the genetic potential of microorganisms, especially actinomycetes, regarding the synthesis of biologically active compounds. Using a variety of algorithms and programs, we predict genes, determine their structure, and predict the functions they perform. In the case of actinomycetes, genome annotation allows the identification of genes encoding enzymes responsible for the synthesis of secondary compounds such as antibiotics, antifungals, anticancer agents, etc. Using methods of structural analysis, it is possible to predict the structure and physicochemical properties of synthesized secondary compounds. This makes it possible to evaluate their potential biological activity and determine possible applications.
Currently, our team has published or is preparing to publish 7 genomes of representatives of the genus Streptomyces, 12 of the genus Umezawaea and 1 genus Mumia.